Next generation sequencing (NGS) refers to advanced DNA sequencing methods that enable sequencing of multiple sequences at the same time. Therefore it is often called as massively parallel sequencing. Originally, the term refers methods that determine sequences of a massively large number (thousands, millions, or more) of short DNA sequences, ranging 100~300 bp simultaneously, Recently, methods that enable sequencing of a very long (even millions of bases) single DNA molecule is belonged to NGS.
Short read sequencing (shotgun sequencing)
In short read sequencing, DNA molecules are fragmented into small pieces, and DNA molecules with specific length is selected, by separating DNA by length using gel electrophoresis. The short pieces of DNAs are subjected to sequencing methods. Usually, primers or adapters were attached at the ends of the fragment. The adapter can bind on the surface of a solid or a bead to segregate each DNA molecules from others. Then the single DNA molecule is amplified by PCR to increase the amount of DNA and to increase sequencing signal. However, the amplication also increase the change of creating technical mutations. The amplified DNA molecules are sequenced using DNA polymerase and dNTPs. Polymerization of different nucleotides transmits different signals, and therefore the sequence of each DNA can be determined.
To summarize, short read sequencing is consisted of the following procedures.
- Fragmentation of DNA
- Sieve DNA by length
- Segregation of single DNA (spatial segregation)
- Amplification of DNA (in segregated region)
- Sequencing of each DNA
Each of the step can be implemented in different ways. Yet, segregation (step 3), amplification (step 4), and sequencing (step 5) methods characterize each sequencing method.
Illumina sequencing
In case of Illumina sequencing, DNAs are placed on the surface of a solid where sequences complementary to the adapter sequences were placed. Adapter of a short DNA molecule binds onto the surface DNA by chance. Each DNA molecule would be scattered over the surface of the solid. Then the sequence can be multiplied by PCR reaction on the surface. During the process, DNA molecules bend and the adapter on the other size will interact to the surface DNA molecule to form a bridge.
The nucleotides can be determined by a series of polymerazation reactions, starting with the primers and dNTPs. A fluorecence dye is placed at the 3′ end of a dNTP, and the color of the dye differes by the base of dNTP. Therefore, we can tell which base is attached at the end of the DNA. The fluorecence dye can be removed to procede polymerization reaction. The next round of polymerazation takes place and the base of the next location can be determined. Repitition of the cycles enable sequencing of a DNA molecule. The reactions take place on the surface of the flow cell, and therefore massively large number of DNA molecules can be sequenced at the same time.
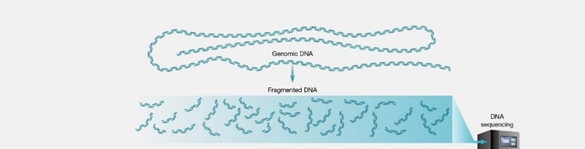
1. Fragmentation
- Fragment DNA into short (100~300 bp) fragments
- Short DNA fragments can be generated by using PCR (eg. 16S amplicon sequencing)
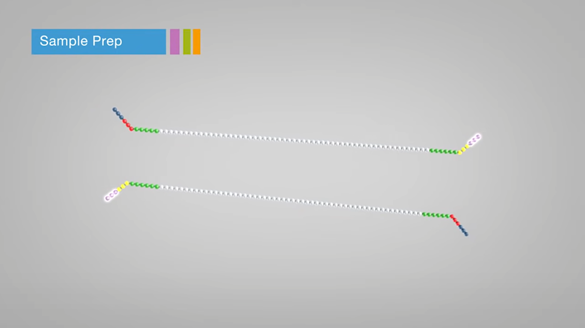
2. Adapter ligation
- Adapter: short DNA fragment complementary to those on the flow-cell
- Bar-codes: sequence tag for samples
- Multiple samples are sequence at a time
- Each sample should be separately tagged by DNA bar-codes
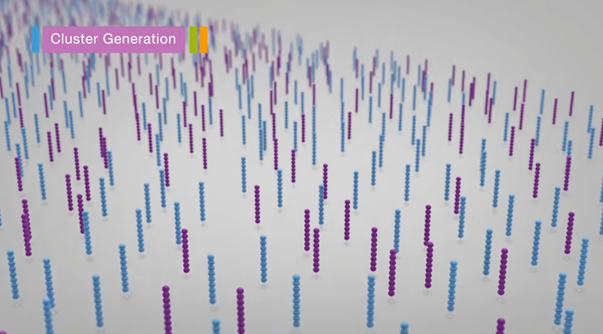
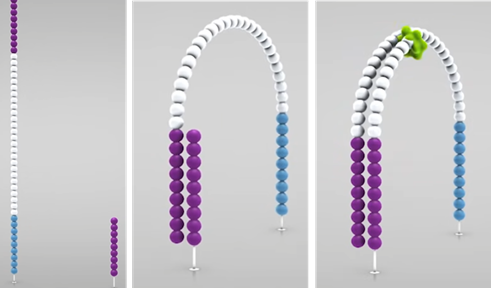
3. Amplification (Bridge PCR)
- Denaturation (dsDNA –> ssDNA)
- Annealing
- Bridge formation
- PCR
4. Sequencing
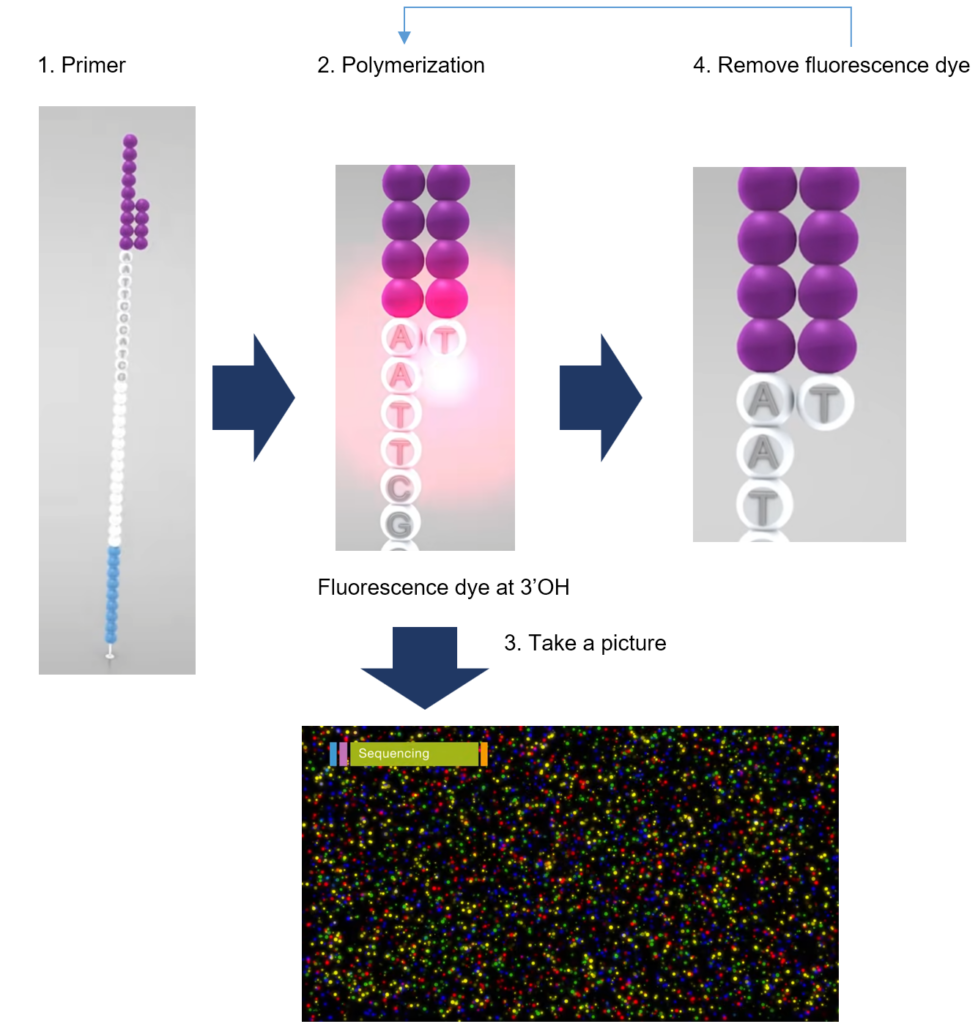
- Primer binding
- Polymerization
- dNTP with reversible terminator
- fluorescence dye at 3’OH
- Take a picture
- Location: individual DNA fragment
- Color: types of the nucleotide
- Time (cycle): position of the nucleotide in the DNA fragment
- Remove fluorescence dye
- Repeat the cycle
Long read sequencing
Oxford nanopore
Applications
Whole genome sequencing and variant detection
